Request Your Free CleanNGS Sample – NGS Cleanup & Size Selection Made Easy
A magnetic bead-based solution for DNA & RNA purification
CleanNGS is your all-in-one reagent for fast, cost-effective purification and double-sided size selection of DNA and RNA. Trusted by researchers worldwide, CleanNGS replaces AMPure XP, SPRI Select, and RNA Clean XP – all in one bottle.
- Compatible with Illumina, Nanopore & automation
- No centrifugation, no columns – just magnetic beads
- Excellent yield, high reproducibility
- Ambient storage – no cold chain needed
- Available for research and CE-IVD clinical workflows
“CleanNGS gave higher DNA yields and consistent results.” – Chris K.
“Cost-effective and easy to automate.” – Michelle M.
Request your free sample here!
Proven Performance: CleanNGS matches golden standard
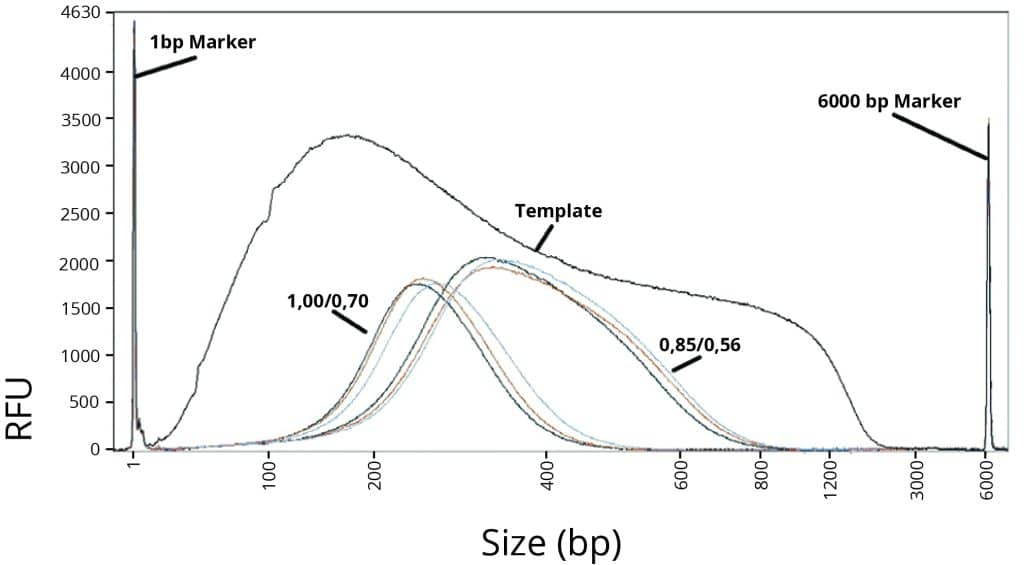
Two different double-size selections (1,00/0,70 and 0,85/0,56) were performed on sheared human genomic DNA. We compared CleanNGS, AMPure XP and SPRI Select, see Figure 1. The results show that CleanNGS performs identical to AMPure XP as well as the SPRI beads. CleanNGS provides consistent double-size selection results and efficiently excludes fragments above and below the target cutoff regions.
Figure 1.
Average DNA fragment size after double-size selection.

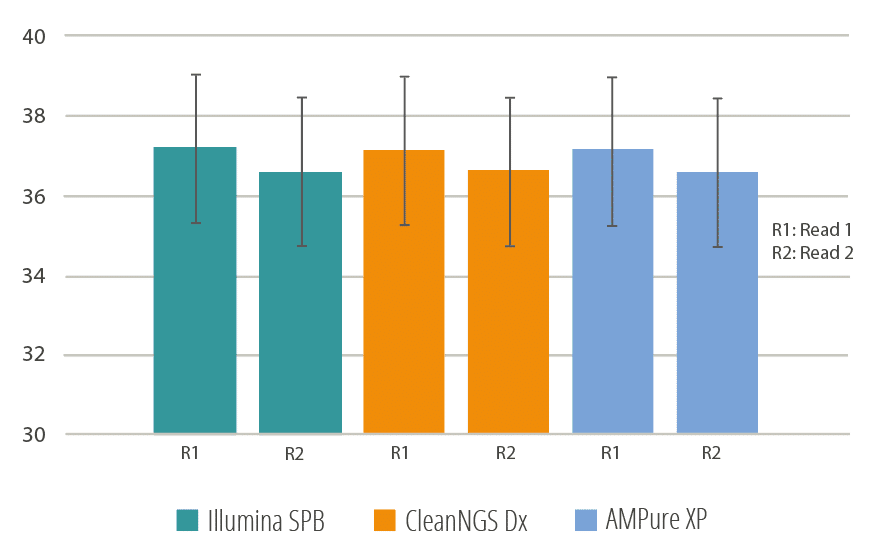
In another experiment, CleanNGS, AMPure XP and Illumina SPB beads were used for purification and double-size selection in a Nextera DNA Flex Library Prep. The generated DNA libraries were sequenced using an Illumina MiSeq and the quality scores of the Read 1 and Read 2 traces were determined. CleanNGS demonstrated equal performance versus AMPure XP and Illumina SPB.
Figure 2.
Average Sequencing Quality score determined of both Read 1 and Read 2 on an Illumina MiSeq instrument.

Amazing results!
These beads are easy to use, and are extremely effective in our applications. The beads produce high quality results and also produce extremely reproducible work. My research would not be the same without these cost-effective and efficient beads. I also appreciate how friendly and the willingness to help that all of the sales team has at GC Biotech to help provide me with what I want.
Very happy and will keep using them.
Some members of the lab were not keen to change away from the regular bead clean up supplier. We tested these and found they gave slightly higher amount of DNA and just as clean.
Like-for-like replacement to the large name-brand equivalents.
Delivers reproducible DNA size-selection for a variety of NGS applications and can be used in with-bead applications.
What does a sample preparation workflow with CleanNGS look like?
CleanNGS is there for you, every step of the way.
To remove reaction buffers or unwanted small fragments, an optional cleanup with CleanNGS can be performed.
Enzymes, salts, and unwanted reaction byproducts are removed in a CleanNGS cleanup step.
A final CleanNGS purification step removes PCR byproducts, such a primer-dimers.
Request your free sample here!
| Order info | ||
|---|---|---|
| Product | Preps | Part number |
| CleanNGS 1 mL | 55 | CNGS-0001 |
| CleanNGS 50 mL | 2,777 | CNGS-0050 |
| CleanNGS 500 mL | 27,777 | CNGS-0500 |
| Product | Preps | Part number |
|---|---|---|
| CleanNGS Dx 50 mL | 2,777 | CNGSDx-0050 |
| CleanNGS Dx 500 mL | 27,777 | CNGSDx-0500 |

