Plant research plays a crucial role in addressing global challenges in agriculture and climate change. By studying plant DNA, scientists can develop more resilient and stronger crops that can withstand diseases, drought and heat. DNA-based screening allows plant breeders to identify desirable traits more efficiently, accelerating the breeding of improved plant varieties. A key step in plant DNA research is DNA extraction. To support this, we offer our Clean Plant DNA Kit.
Our Clean Plant DNA Kit includes optimised lysis and wash buffers for leaves and seeds for a variety of plant species. Together with the Plant DNA Particle Solution, the kit delivers pure DNA free from inhibitors such as polysaccharides and phenolic compounds. Extracted DNA is of sufficient purity to use in downstream detection procedures based on NGS and PCR analysis, whilst the protocol can easily be integrated into both manual and workflows as well as automated workflows, such as on our CleanXtract 96.



Plant DNA research has revolutionized agriculture, environmental science, and biotechnology. Downstream applications in these fields include Next-Generation Sequencing (WGS, RNA-Seq), (q)PCR, KASP, SNP genotyping and CRISPR/Cas9 gene editing.
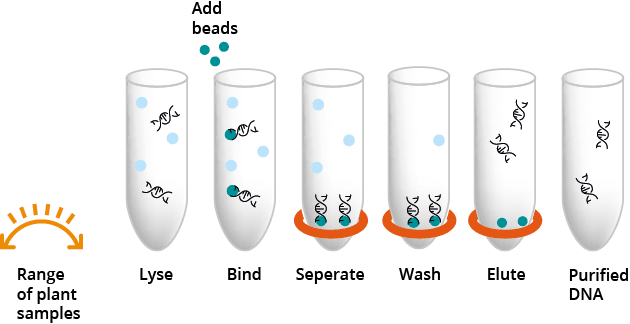
Plant samples are first homogenised and lysed with the included lysis buffer. Plant DNA Particle Solution is added to the lysed plant material and the plant DNA binds to the surface of the magnetic particles. The magnetic particles are separated from the lysate by a magnetic separation device. After the required wash buffer and ethanol wash steps, the purified DNA is eluted with the elution buffer.
DNA Integrity
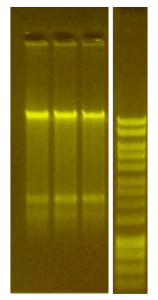
To check the integrity of extracted genomic DNA, the Clean Plant DNA Kit was used to extract DNA from maize seeds. Input material was 1 seed per sample and elution was performed in 60 µL Elution Buffer. The size distribution of the extracted DNA visualized on an agarose gel indicates high genomic DNA integrity.
Figure 1: Agarose gel (0,7%) electrophoresis analysis of genomic DNA extracted from maize seeds (10 μL sample per slot). Lanes 1-3 contain DNA from maize seeds and lane 4 contains a 1kb ladder.
DNA Purity
In addition to the agarose gel analysis, OD measurements were performed on extracted maize samples (1 maize seed input and 60 µL elution) to show the purity of the DNA extracted from maize seeds. The 260/230 ratio greater than 2,0 and 260/280 ratio greater than 1,8 indicate that the extracted DNA is free of protein, polyphenols or other contaminants, making it suitable for all downstream applications indicated (Table 1).
TABLE 1. OD measurements of eluted DNA showing DNA concentration and purity from maize seeds.
| n=72 | ng/μL ± SD | 260/230 ± SD | 260/280 ± SD |
| Maize | 366 ± 28 | 2,06 ± 0,10 | 2,03 ± 0,01 |
In another experiment, DNA was extracted from tomato and cucumber leaves. Similar to the OD values of the DNA extracted from plant seeds, the 260/230 ratio greater than 2,0 and 260/280 ratio greater than 1,8 indicating that the DNA extracted from leaves is free of contaminants and suitable for all downstream applications indicated (Table 2).
TABLE 2. OD measurements of eluted DNA comparing DNA concentration and purity from tomato and cucumber leaves.
| n=10 | ng/μL ± SD | 260/230 ± SD | 260/280 ± SD |
| Tomato | 37 ± 3 | 2,14 ± 0,05 | 2,02 ± 0,06 |
| Cucumber | 50 ± 3 | 2,38 ± 0,04 | 2,02 ± 0,03 |
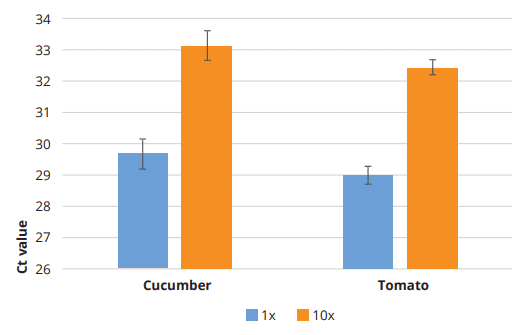
To confirm that the Clean Plant DNA Kit is compatible with downstream applications like qPCR, an extraction from 6 cucumber leave samples and 8 tomato leave samples spiked with an internal extraction control (Meridian Bioscience) was performed. qPCR on the undiluted and 10x diluted eluates targeting the internal extraction control DNA exhibited an average Ct value of 29,69 ± 0,47 for cucumber and 29,00 ± 0,29 for tomato, which was in the expected range for the control DNA used (29 ± 1,6). Between the 1x and 10x dilutions there is a Ct value difference of 3,47 for cucumber and 3,46 for tomato. These results confirm that there is no inhibition of the polymerase reaction due to polyphenols or other plant chemicals and that the amplification steps after extraction were successful utilizing the Clean Plant DNA Kit.
Figure 2: qPCR results of qPCR Extraction Control Red DNA (Meridian Bioscience) after a Clean Plant DNA Kit extraction procedure from cucumber and tomato leaves (1x and 10x dilution).
List of Analyzed materials:
• Barley*
• Blackberry
• Brassica
• Carnation
• Chrysantum
• Cornsalad
• Cucumber
• Endive
• Kalanchoe
• Maize*
• Melon
• Onion
• Potato
• Rapeseed*
• Raspberry
• Rose
• Soy*
• Spinach
• Squash
• Strawberry
• Sunflower*
• Tomato
• Wheat*
*DNA extracted from seeds
There are no reviews yet. Be the first one to write one.