Next Generation Sequencing (NGS) has revolutionized molecular diagnostics with its capacity of massive parallel sequencing of DNA or RNA fragments to detect clinically important genomic variants and mutations. A critical step in generating high-quality NGS data is the preparation of pure DNA or RNA of specific lengths. To support this, we offer CleanNGS Dx for library cleanup and size selection, ensuring the highest levels of precision and consistency in your diagnostic workflows.
Our special buffer formula ensures optimal size selection for NGS libraries, and the high-quality magnetic beads allow faster separations and better RNA/DNA recovery.
CleanNGS Dx is produced RNase free, which makes it an ideal solution for all downstream RNA or DNA NGS experiments.



CleanNGS Dx is suited for cleanup in between NGS library preparation steps, or post library preparation for size selection, in diagnostic workflows.
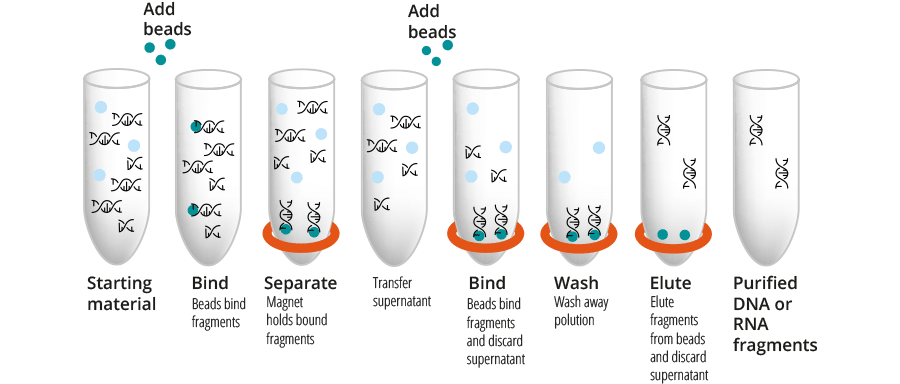
For double sided size selection, we first add CleanNGS Dx reagent with magnetic beads in a certain volume ratio. Separate the large DNA or RNA fragments from the solution with a magnetic plate and add more CleanNGS Dx reagent to the supernatant to clean up the small DNA fragments and inhibitors. After two washing steps, the purified DNA or RNA is eluted.
Figure 1
Average DNA fragment size after double-size selection using various CleanNGS Dx ratios.
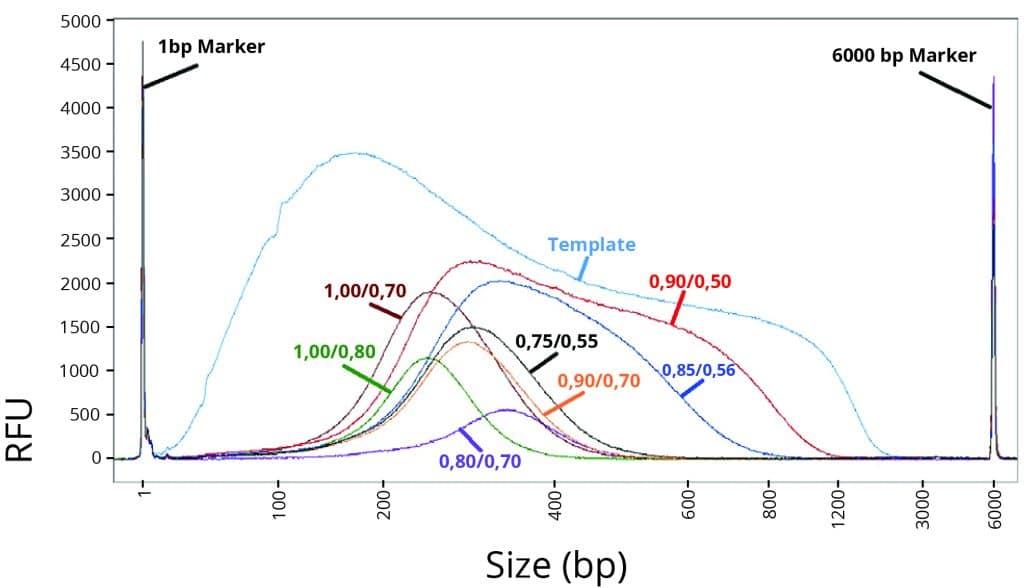
Figure 1 shows an overview of CleanNGS Dx size selection capabilities by alteration of the CleanNGS Dx versus sample volume ratio.
Figure 2
Average DNA fragment size after double-size selection.
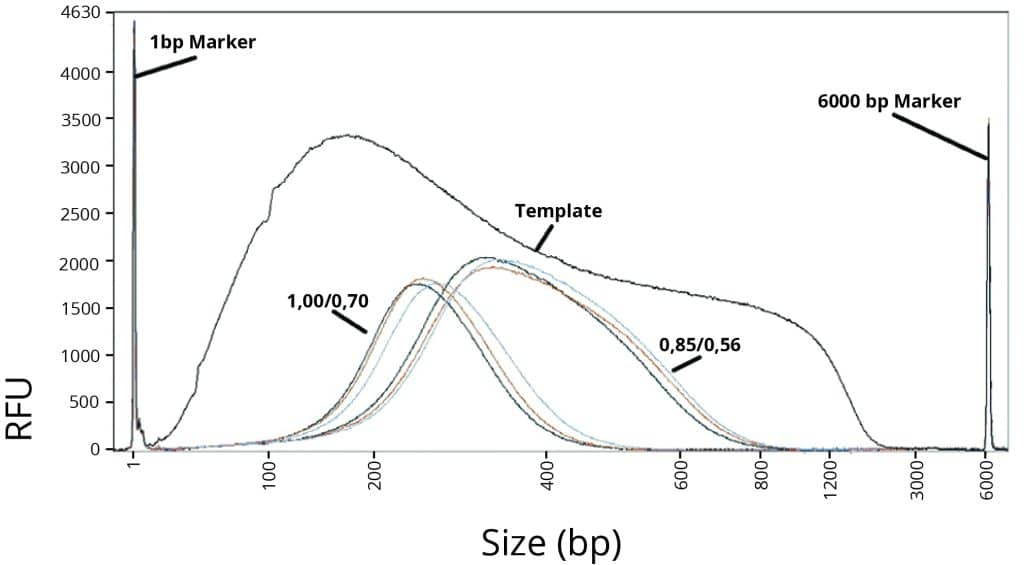
In Figure 2, two different double-size selections (1,00/0,70 and 0,85/0,56) were performed on sheared human genomic gDNA to compare CleanNGS Dx, Competitor A and Competitor S. The results show CleanNGS Dx performs identical to Competitor A as well as Competitor S. This demonstrates that CleanNGS Dx provides consistent size selection results and efficiently excludes fragments above and below the target cutoff regions.
Analyses for Figure’s 1 and 2 were conducted on the Agilent Fragment Analyzer 5200, using the HS NGS Fragment Kit.
Figure 3
Average Sequencing Quality score determined of both Read 1 and Read 2 on all Illumina MiSeq instrument.
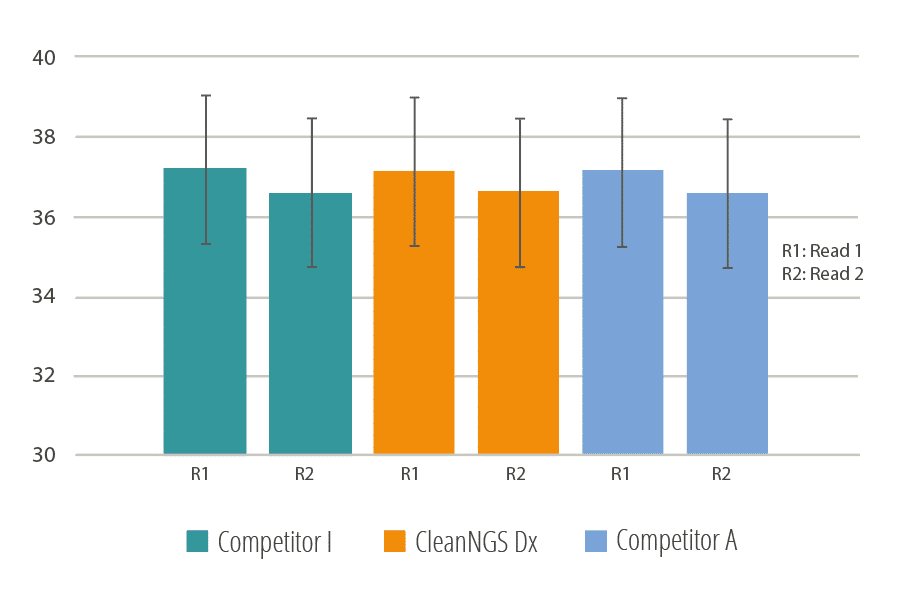
In Figure 3 CleanNGS Dx and two other commercially available kits (I and A) were used for purification and double-size selection in a Nextera DNA Flex Library Prep. The generated DNA libraries were sequenced using an Illumina MiSeq and the quality scores of the Read 1 and Read 2 traces were determined. CleanNGS Dx demonstrated equal performance versus the competitor products.
Figure 4
Recovery percentage after single-size selection purification determined by dsDNA fluorescence assay. (N=48)
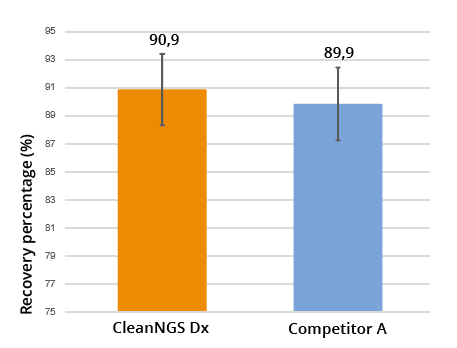
Figure 4 compares percentage recovery of CleanNGS Dx compared to competitor A. 50μL of sheared genomic DNA was purified using a 1.8x ratio and the recovery was then determined versus the unpurified sample material. CleanNGS Dx exhibits an outstanding recovery above 90%.
Figure 5
RNA Quality Number determined after purification of total RNA, using the Agilent fragment analyzer.
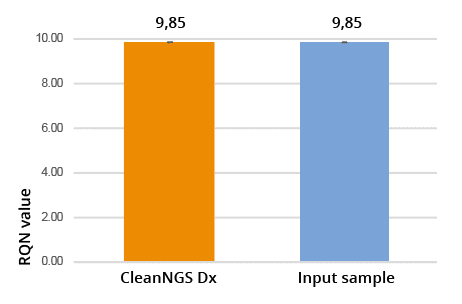
In Figure 5, 10uL of total RNA control was purified using CleanNGS Dx using a 1.8x sample volume ratio with the recovered RNA compared to the unpurified total RNA control. The RQN values of the purified versus the unpurified samples indicate no degradation of the RNA during purification, clearly demonstrating that CleanNGS Dx delivers superior RNA quality for downstream applications.
There are no reviews yet. Be the first one to write one.
CleanNA
Coenecoop 75
2741 PH Waddinxveen
The Netherlands
V.A.T. – NL859255955B01
KVK – 72839511
T: +31 (0) 182 22 33 50
E: info@cleanna.com
Technical support
E: support@cleanna.com